Part 1: DeepGauge's coverage critiera analysis results: MNIST
On MNIST dataset, we have studied three LeNet family models, including LeNet-1, LeNet-4, and LeNet-5) to analyze our criteria. Our proposed neuron-level coverage criteria needs to have the information (i.e., UpperBound, LowerBound) of each neuron. The whole DNN can be considered as the one that is programmed by training data that decides main functional region and corner-case region of each neuron. To obtain such information, we profile the DNNs runtime execution results (outputs) of each neuron using all the 60,000 training data of MNIST. For each DL model under analysis, we run the 10,000 test data on the model to obtain the different coverage cases. For each studied DL model, we also generate another four sets of adversarial test data, through three well-known adversarial techniques i.e., FGSM, BIM, JSMA, and CW.
After generating the three adversarial datasets, we aggregate each of them with the original MNIST test dataset (with a total size 20,000 for each), which enables us to perform the comparative study on the effectiveness of the existing MNIST test dataset and how adversarial test data enhances the defect detection ability, from our coverage criteria measurement.
Our paper shows a set of running configurations for evaluation. To be more thorough, this website gives all
the experimental settings we have performed. For each criterion, we also evaluate the following parameters,
in particular, we have in total 3(models) * 5 (datasets) * 14(criteria configurations) = 210 configurations
The 5 datasets for each model are consisted of one original test data, and 4 adversarial generated data by FGSM, BIM, JSMA and cW (aggregated with original test data), respectively. It is worth noting that these adversarial data generation is often model dependent, the adversarial datasets are actually different for each model, although the number of datasets to evaluate on a model is 4. For example, for FGSM algorithm, we actually use it generate three adversarial datasets, one for each of LeNet-1, LeNet-4, and LeNet-5, respectively.
The detailed configurations for each criterion are shown as follows:
We give the detailed discussion and explanation on the MNIST resulting data, and highlight the difference for results obtained on ImageNet. For simplicity, all Figures in section 1 (MINIST) would be considered as Figure 1 set with subfigure (a), (b), (c), etc.; similarly, all Figures in section 2 (ImageNet) would be the Figure 2 set.
Coverage result plot for each critera and configurations:
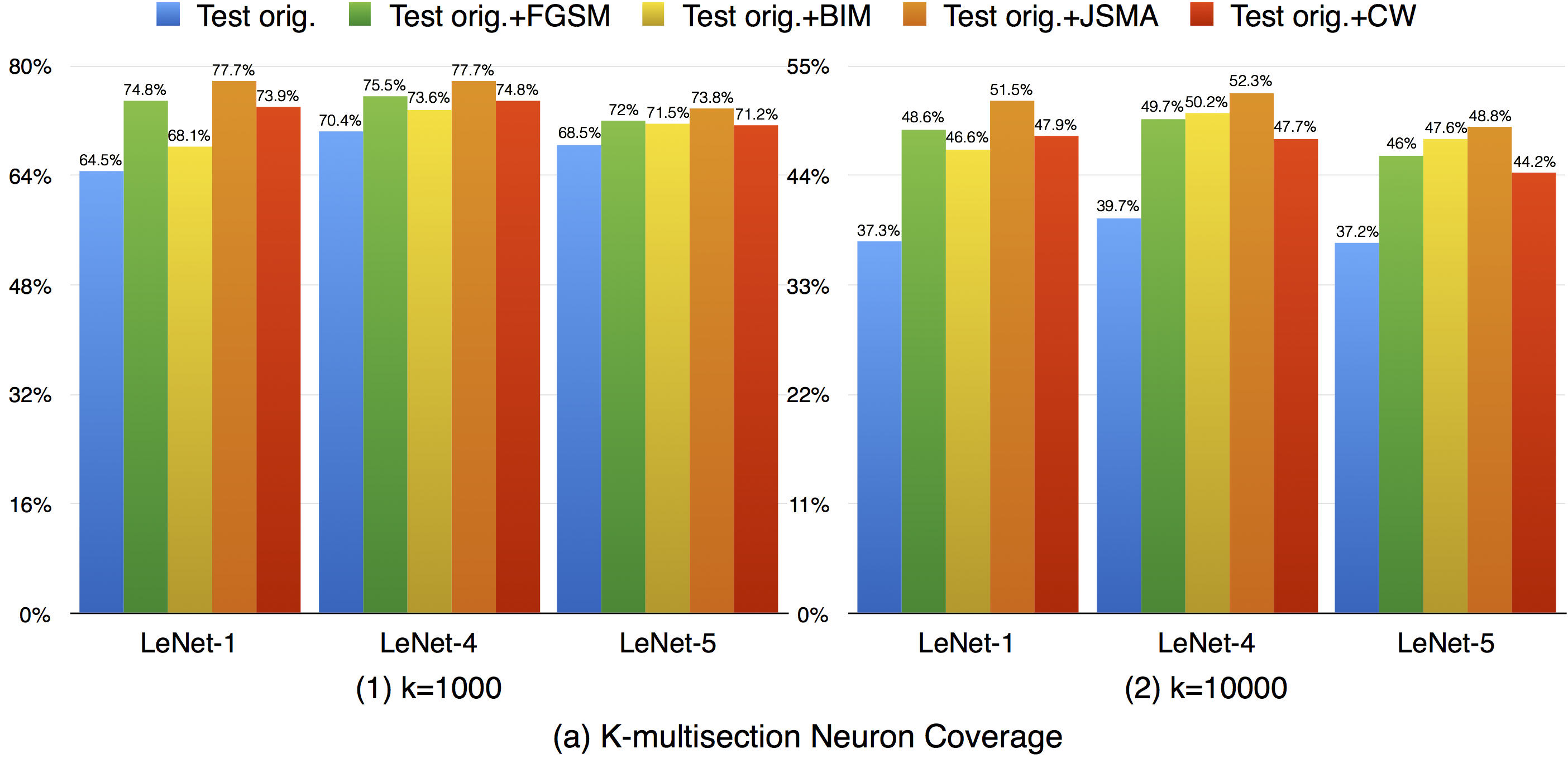
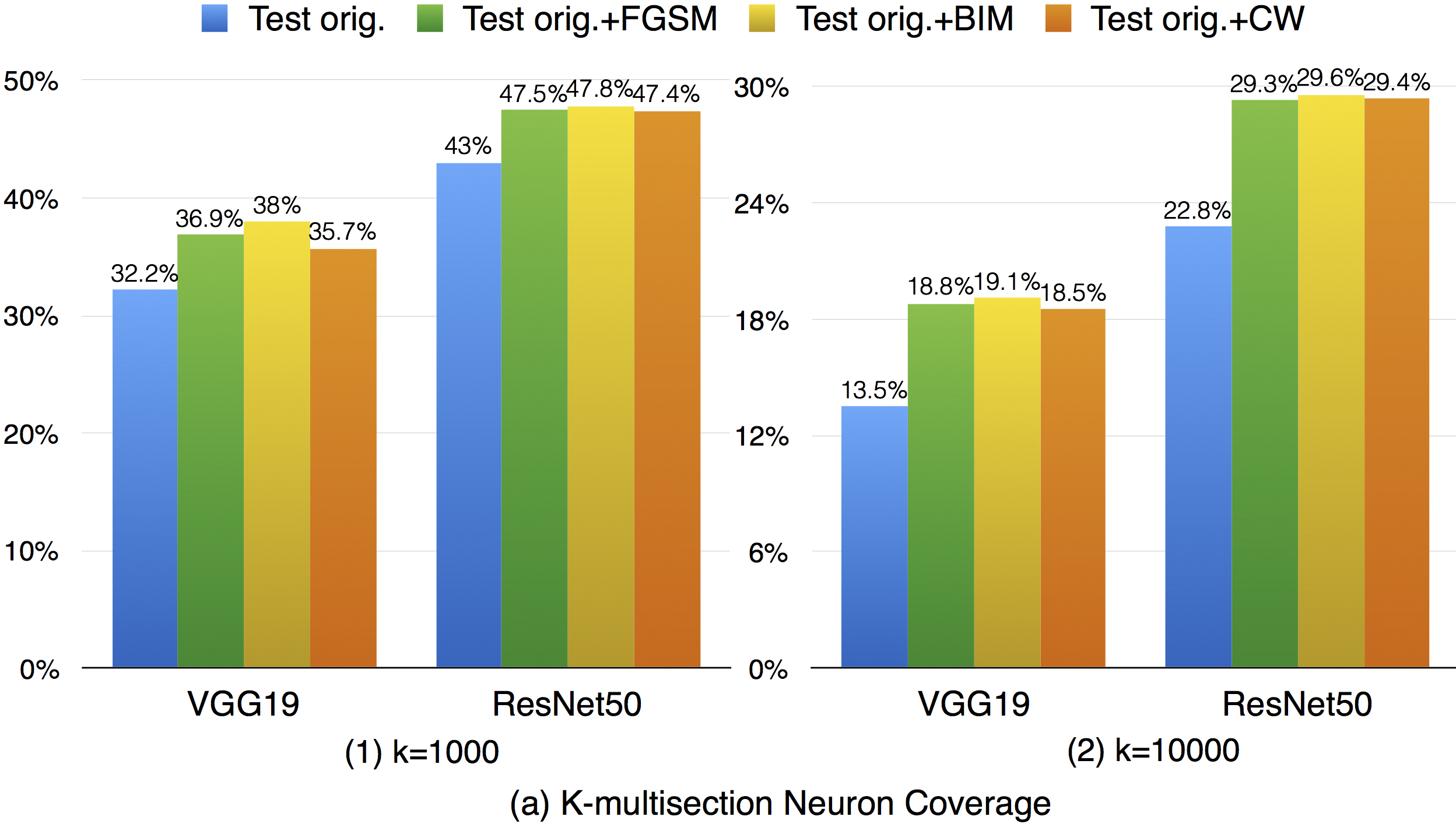
For k-multisection neuron coverage, we performed on two settings, where k=1,000, and k=10,000, which are reasonable choice considering the test dataset size. On these two settings, we find that the adversarial test data could cover different main functional region compared with original test data, which could be observed in both settings, where the coverage improvement for k=10,000 setting is more obvious compared with k=1,000 due to is finer granularity. This means that increasing the k-multisection neuron coverage could potentially detect more defects of a DNN. On the all evaluated MNIST configurations, we find that JSMA shows slightly better performance to obtain higher k-multi-section coverage compared with other FGSM and BIM. Although these adversarial techniques improve this coverage compared with original test data, there are still many main functionality that are missed by the adversarial data.
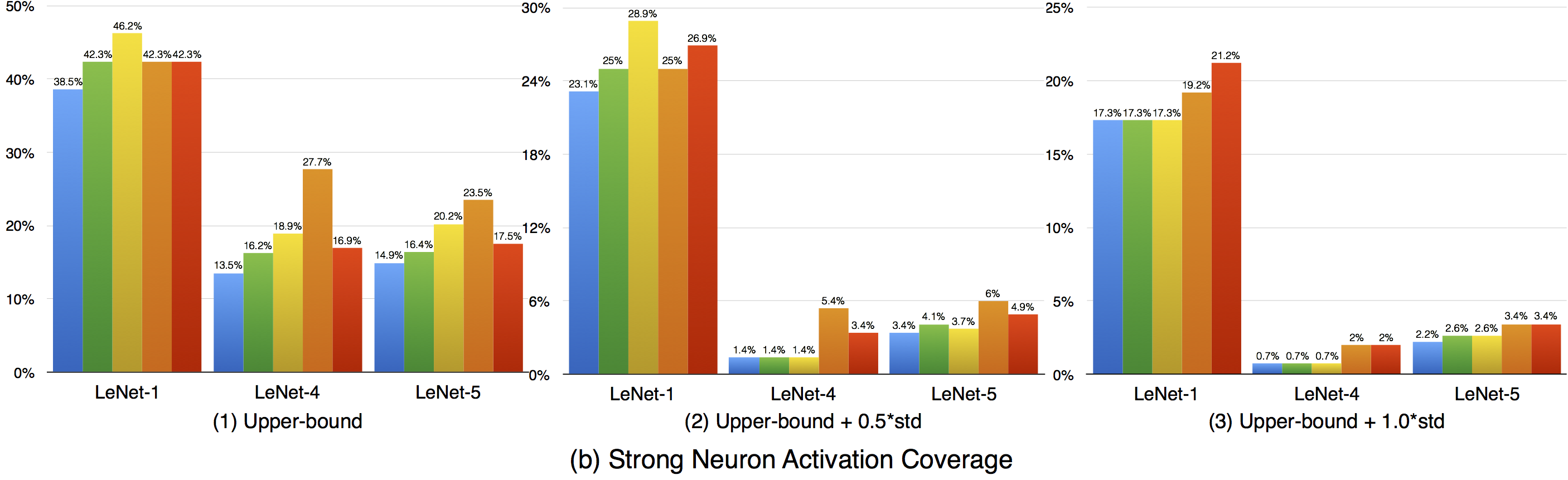
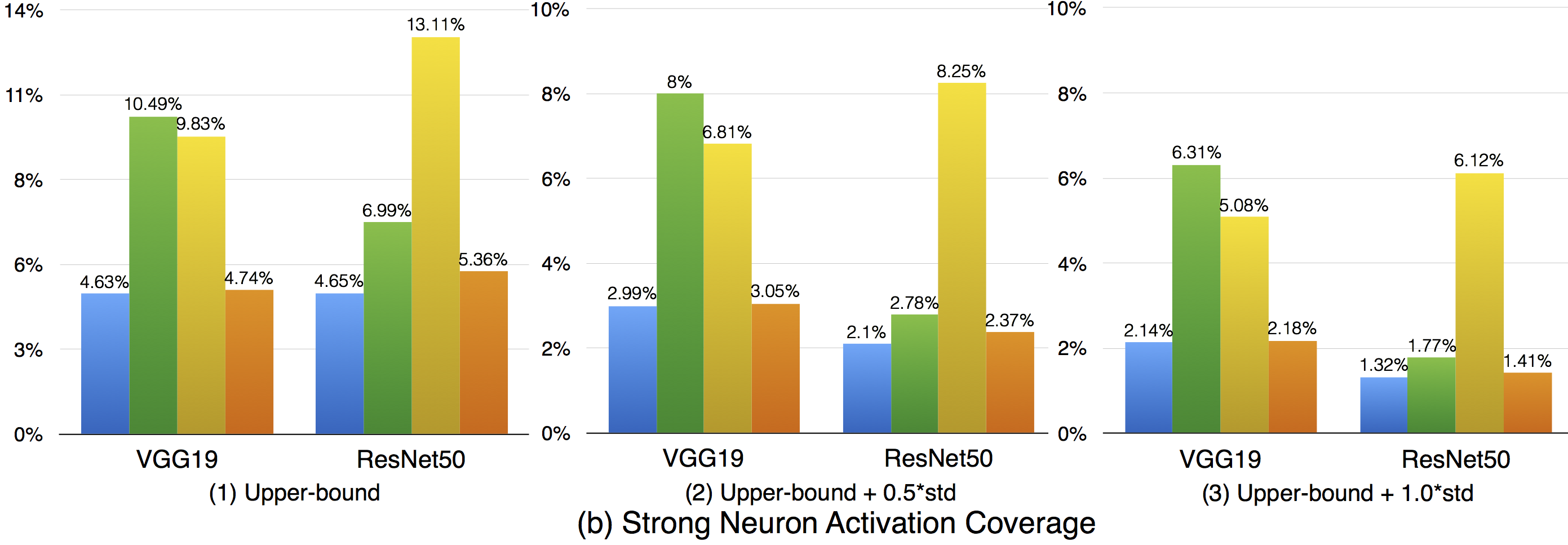
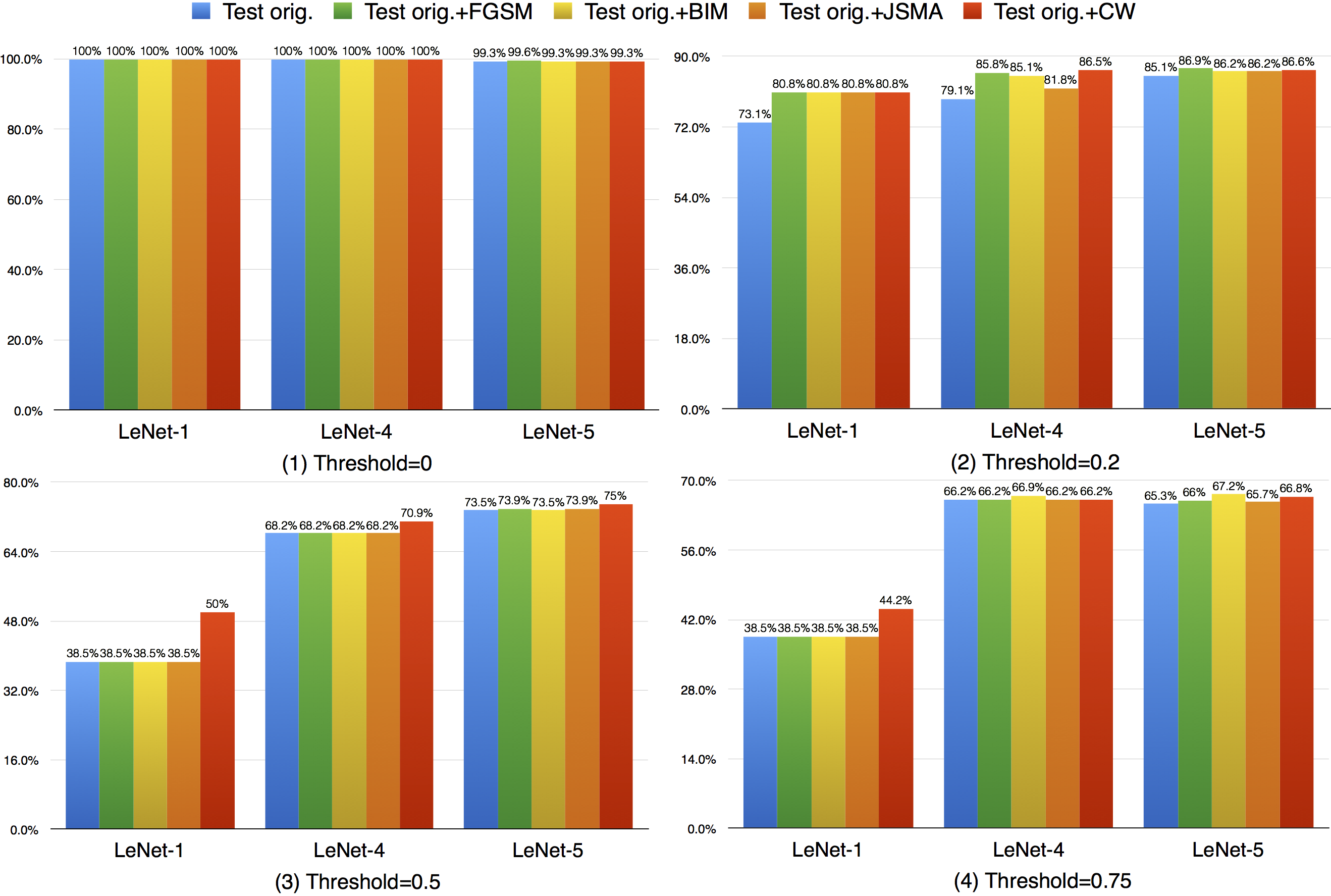
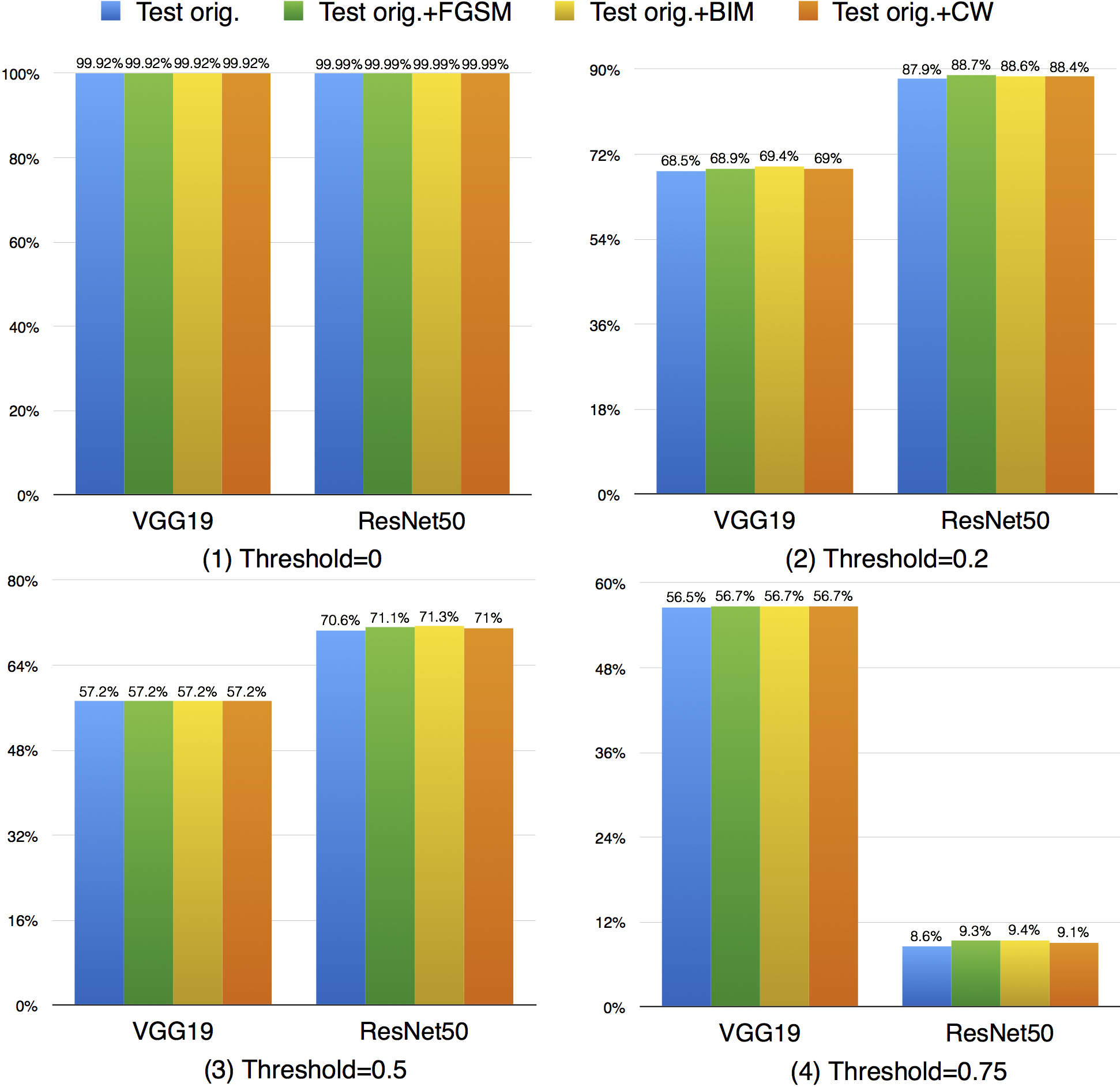
Compared with k-multisection neuron coverage, strong neuron activation coverage achieved for original test data is generally lower on each evaluated configuration, indicating that original test data focuses more on testing the main functionality of DNNs. Note that, we use three UpperBound settings, the larger its value, the higher deviation (more rare cases) from the main functional region. The resulting data is consistent with these 3 settings, the larger Upperbound is given (e.g., UpperBound+std), it is more difficult to cover the target region, resulting in lower coverage. Although adversarial data could greatly improve the strong neuron activation coverage (e.g., for LeNet-4, JSMA improves the coverage by 105%, from 0.135 to 0.277 for configuration 1; by 286%, from 0.014 to 0.054 for configuration 2; and by 186%, from 0.007 to 0.02, for configuration 3), the total coverage obtained is still low, and it is still necessary to design other test data generation techniques that could effectively cover more on these missed regions.
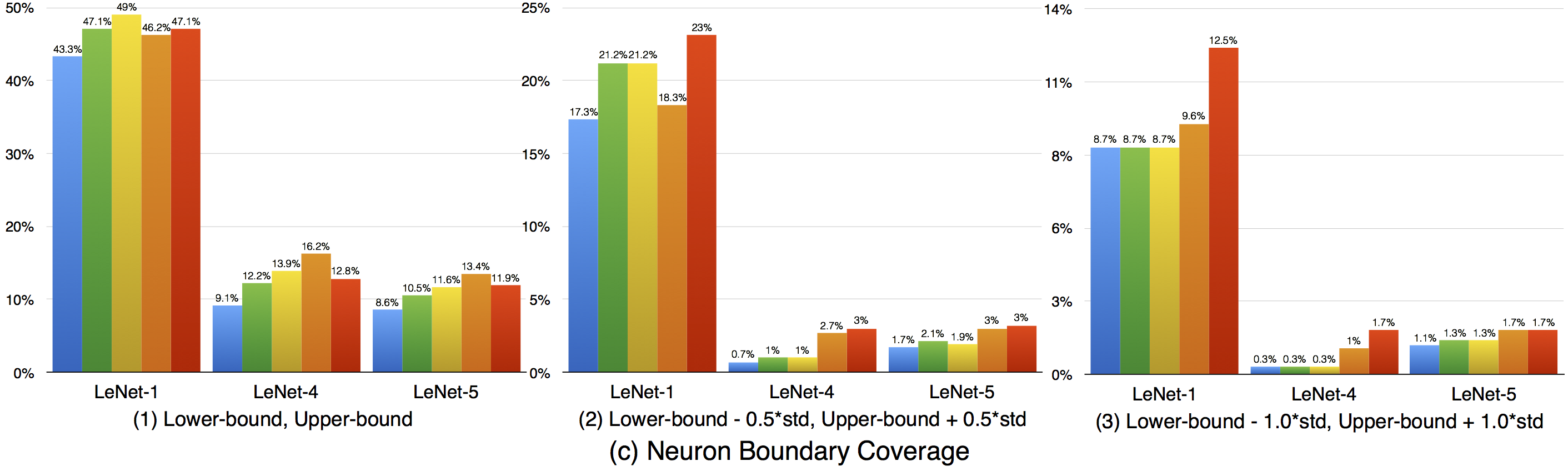
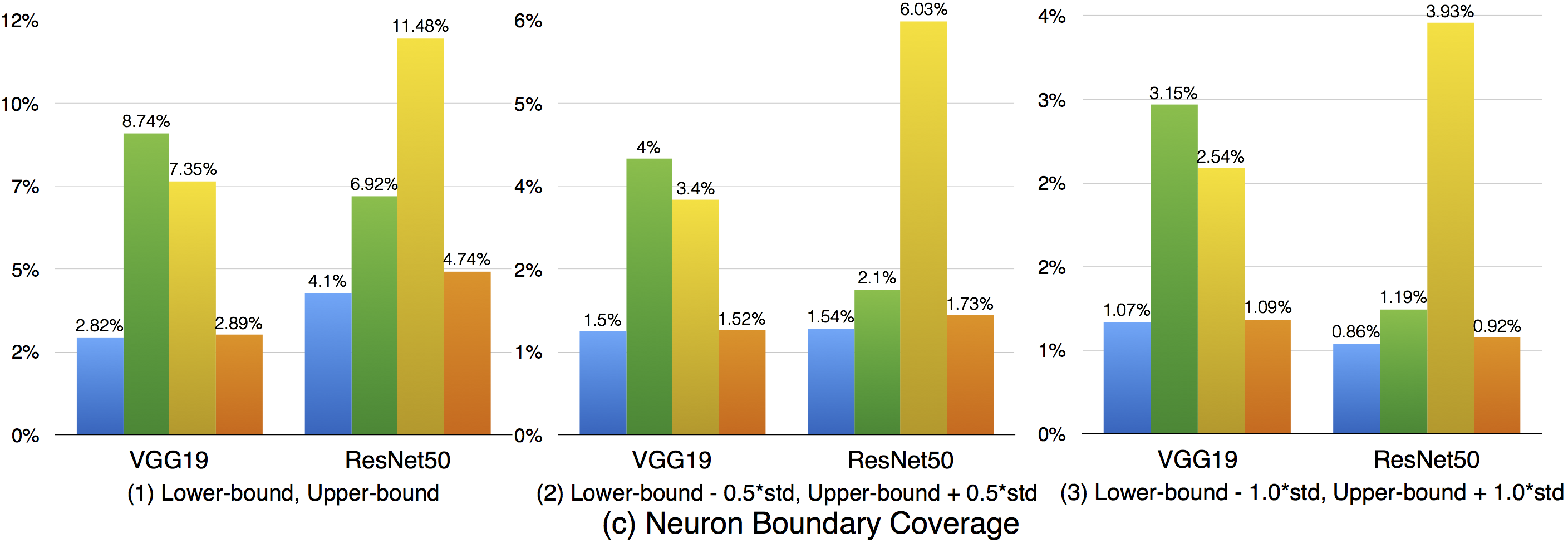
Compared with strong neuron activation coverage that focuses on the neuron strong activation boundary case, the neuron boundary coverage also takes the non-activated boundary cases into consideration. Our empirical observation shows that the neuron non-activation cases also play an important role in final decision calculation (e.g., classification, prediction). In line with the strong neuron activation coverage on each corresponding configuration, we can see that neuron boundary coverage are often lower in most cases (LeNet-1 is an exception, which might be caused by its small neural network size), indicating that the covering the non-activated boundary on studied DNN could be even harder than covering the strong neuron activation case. This might be caused by the intrinsicality of the activation function used in the DNN, where ReLu would block the all negative cases and set output to zero. Since the adversarial generated test data would trigger the defects of DNN, it means that defects of DNNs could also exist in the corner-case regions, and further adversarial test data generation technique design should take this into account, which is missed by current techniques.
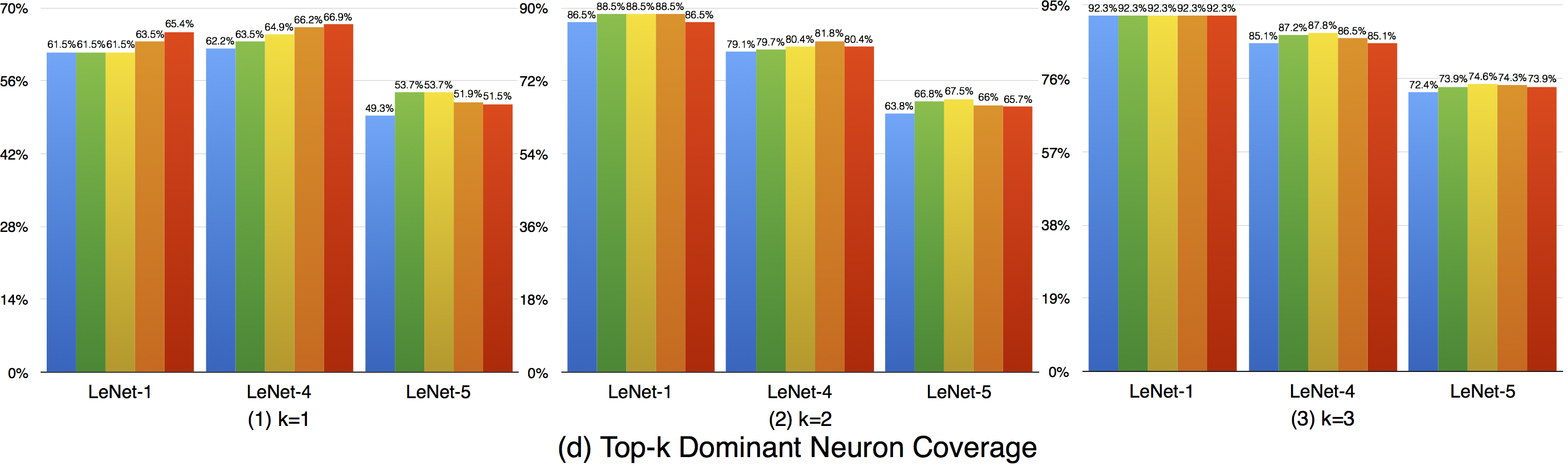
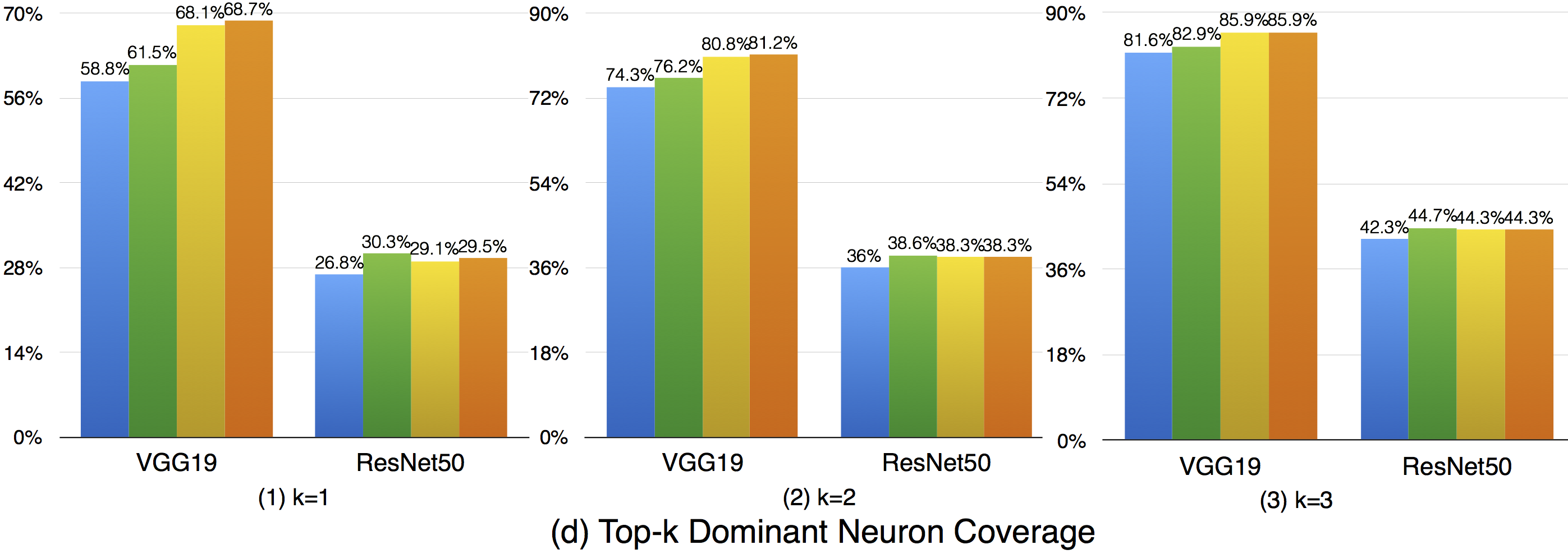
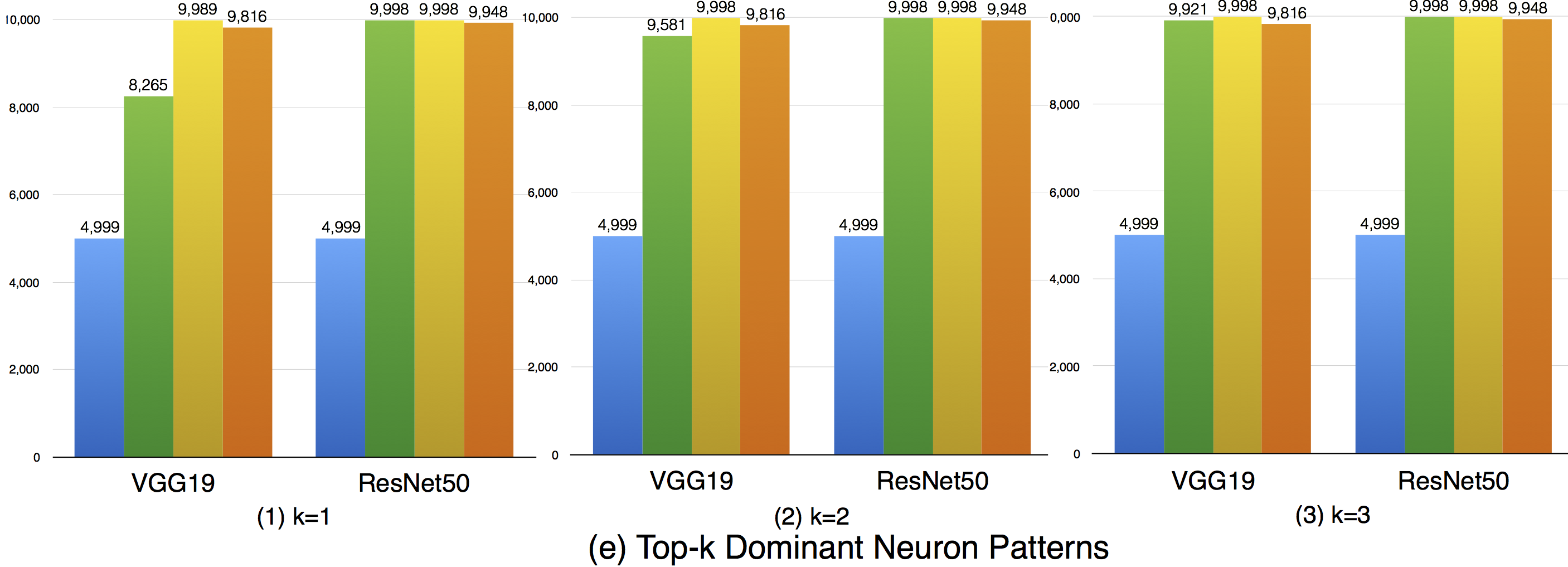
Top-k dominant neuron coverage is a layer-level coverage criterion. For an input data, if a neuron' output is among the top k most activated neurons of a layer, it is defined as covered. In other words, this criterion measures whether a neuron has ever played a dominant role in its layer among all the test data.
In our evaluation, we used three configurations (i.e., k=1,2,3) to evaluate the top-k dominant neuron coverage and observe the behavior under different cases.
Overall, the adversarial test data does not improve this coverage criterion much compared with the original dataset. This means that the subset of most dominant neurons of each layer tends to be stable. As k increases, higher coverage is obtained, so a neuron of a layer may not ever paly as the most (top-1) dominant neuron, but it still may play as the second dominant or third dominant neurons of a layer. In the top-3 case, the overall coverages are relatively high, indicating that most neurons have played as one of the top-3 dominant neurons to support the function of DNNs.
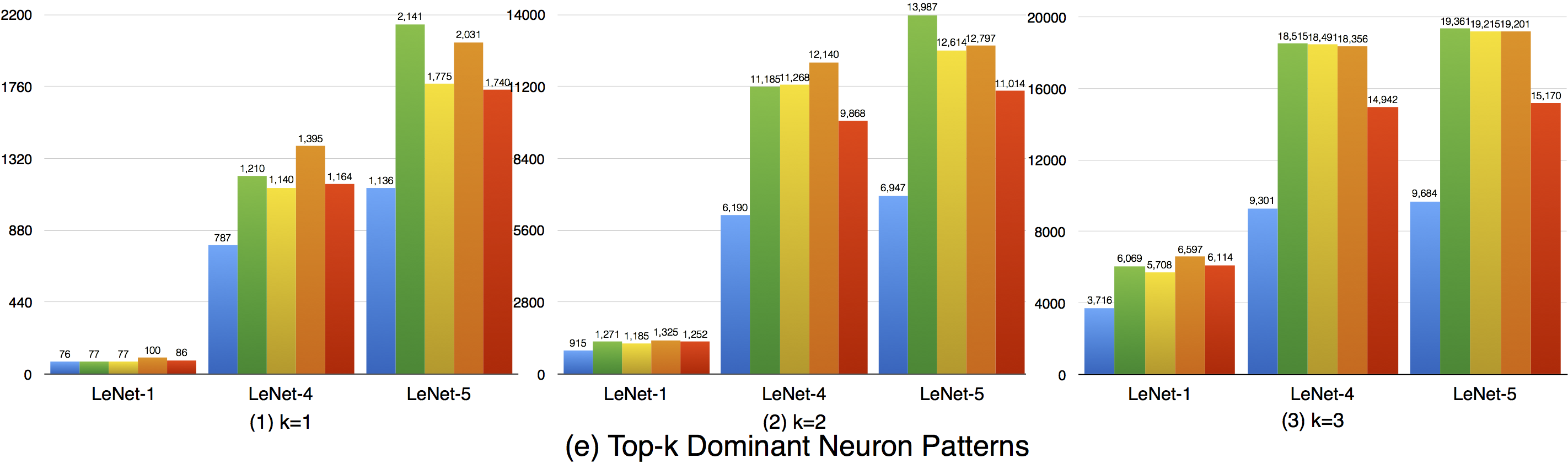
Although the top-k dominant neuron subset of each layer is relatively stable, the top-k dominant neuron patterns are still able to distinguish different inputs in many cases. In particular, as k, DNNs neuron size and complexity grow, more different input data could be uniquely identified (see k=1 to 3).
Furthermore, we could also see obvious dominant neuron pattern improvement for adversarial test data compared with original test data. This means that original test data and adversarial generated test data trigger quite different top-k dominant neuron patterns, and improving this criterion could increase the chance to detect DNN's defects.
In summary, (1) top-k dominant neuron coverage describes the set of major dominant neurons of the DNN during its functioning; (2) although the top-k dominant neuron subset of each layer is relatively stable, their neuron patterns could mostly distinguish the input data; (3) adversarial test data follows different top-k dominant neuron patterns that trigger DNN defects compared with original test data that trigger the correct behavior of DNNs. Therefore, improving the top-k dominant neuron pattern would be an important indicator to increase the possibility for defect detection of DNNs.